Brequinar derivatives and species-specific drug design for dihydroorotate dehydrogenase.
Hurt, D.E., Sutton, A.E., Clardy, J.(2006) Bioorg Med Chem Lett 16: 1610-1615
- PubMed: 16406782
- DOI: https://doi.org/10.1016/j.bmcl.2005.12.029
- Primary Citation of Related Structures:
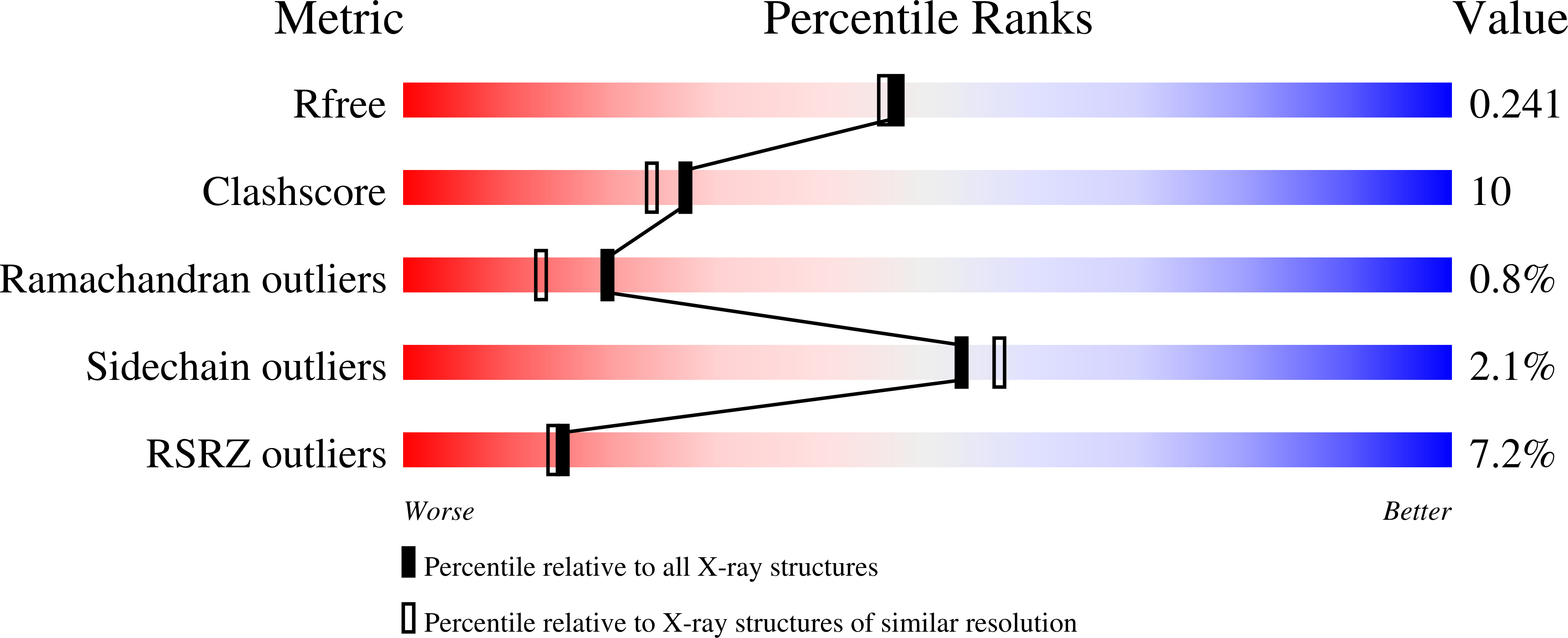
2B0M - PubMed Abstract:
Therapeutic agents brequinar sodium and leflunomide (Arava) work by binding in a hydrophobic tunnel formed by a highly variable N-terminus of family 2 dihydroorotate dehydrogenase (DHODH). The X-ray crystallographic structure of an analog of brequinar bound to human DHODH was determined. In silico screening of a library of compounds suggested another subset of brequinar analogs that do not inhibit human DHODH as potentially effective inhibitors of Plasmodium falciparum DHODH.
Organizational Affiliation:
Department of Chemistry and Chemical Biology, Cornell University, Ithaca, NY 14850, USA.